
Transposition Occurs by Both Replicative and Nonreplicative Mechanisms
 المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
 المصدر:
LEWIN’S GENES XII
المصدر:
LEWIN’S GENES XII
 الجزء والصفحة:
الجزء والصفحة:
 22-4-2021
22-4-2021
 3760
3760
Transposition Occurs by Both Replicative and Nonreplicative Mechanisms
KEY CONCEPTS
- Most transposons use a common mechanism in which staggered nicks are made in target DNA, the transposon is joined to the protruding ends, and the gaps are filled.
- The order of events and exact nature of the connections between transposon and target DNA determine whether transposition is replicative or nonreplicative.
The insertion of a transposon into a new site is illustrated in FIGURE 1. It consists of making staggered breaks in the target DNA, joining the transposon to the protruding single-stranded ends, and filling in the gaps. The generation and filling of the staggered ends explain the occurrence of the direct repeats of target DNA at the site of insertion. The stagger between the cuts on the two strands determines the length of the direct repeats; thus, the target repeat characteristic of each transposon reflects the geometry of the enzyme involved in cutting target DNA.
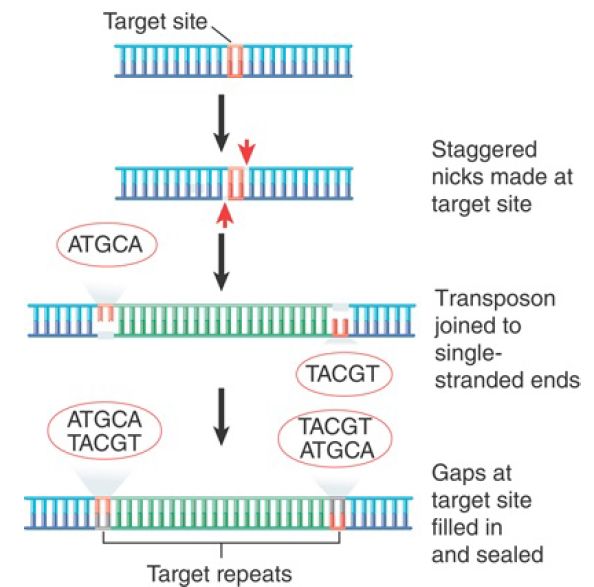
FIGURE 1.The direct repeats of target DNA flanking a transposon are generated by the introduction of staggered cuts whose protruding ends are linked to the transposon.
The use of staggered ends is common to most means of transposition, but we can distinguish two major types of mechanisms by which a transposon moves:
- In replicative transposition, the element is duplicated during the reaction so that the transposing entity is a copy of the original element. FIGURE 2. summarizes the results of such a transposition. The transposon is copied as part of its movement. One copy remains at the original site, whereas the other inserts at the new site. Thus, transposition is accompanied by an increase in the number of copies of the transposon. Replicative transposition involves two types of enzymatic activity: a transposase that acts on the ends of the original transposon and a resolvase that acts on the duplicated copies. Although one group of transposons moves only by replicative transposition , true replicative transposition is relatively rare among transposons in general.
- In nonreplicative transposition, the transposing element moves as a physical entity directly from one site to another and is conserved. The insertion sequences and composite transposons (Tn), Tn10 and Tn5 (as well as many eukaryotic transposons), use the mechanism shown in FIGURE 3, which involves the release of the transposon from the flanking donor DNA during transfer. This type of mechanism, often referred to as “cut-and-paste,” requires only a transposase.
Another mechanism utilizes the connection of donor and target DNA sequences and shares some steps with replicative transposition. Both mechanisms of nonreplicative transposition cause the element to be inserted at the target site and lost from the donor site. What happens to the donor molecule after a nonreplicative transposition? Its survival requires that host repair systems recognize the double-strand break and repair it .
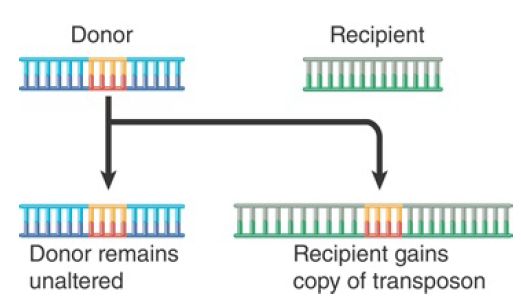
FIGURE 2. Replicative transposition creates a copy of the transposon, which inserts at a recipient site. The donor site remains unchanged, so both donor and recipient have a copy of the transposon.
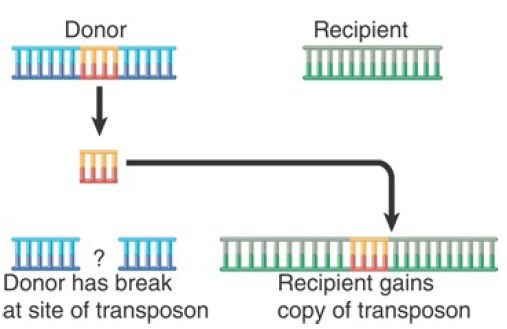
FIGURE 3. Nonreplicative transposition allows a transposon to move as a physical entity from a donor to a recipient site. This leaves a break at the donor site, which is lethal unless it can be repaired.
Some bacterial transposons use only one type of pathway for transposition, whereas others may be able to use multiple pathways. The elements IS1 and IS903 use both nonreplicative and replicative pathways, and the ability of phage Mu to turn to either type of pathway from a common intermediate has been well characterized.
The same basic types of reaction are involved in all classes of transposition events. The ends of the transposon are disconnected from the donor DNA by cleavage reactions that generate 3′–OH ends. The exposed ends are then joined to the target DNA by transfer reactions, involving transesterification in which the 3′–OH end directly attacks the target DNA. These reactions take place within a nucleoprotein complex that contains the necessary enzymes and both ends of the transposon. Transposons differ as to whether the target DNA is recognized before or after the cleavage of the transposon itself, and whether one or both strands at the ends of the transposon are cleaved prior to integration.
The choice of target site is in effect made by the transposase, sometimes in conjunction with accessory proteins. In some cases, the target is chosen virtually at random. In others, there is specificity for a consensus sequence or for some other feature in the target. The feature can take the form of a structure in DNA, such as bent DNA, or a protein–DNA complex. In the latter case,
the nature of the target complex can cause the transposon to insert at specific promoters (such as Ty1 or Ty3, which select pol III promoters in yeast), inactive regions of the chromosome, or replicating DNA.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة


