النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
snRNAs Are Required for Splicing
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
12-5-2021
2213
snRNAs Are Required for Splicing
KEY CONCEPTS
- The five snRNPs involved in splicing are U1, U2, U5, U4,and U6.
- Together with some additional proteins, the snRNPs form the spliceosome.
- All the snRNPs except U6 contain a conserved sequence that binds the Sm proteins that are recognized by antibodies generated in autoimmune disease.
The 5′ and 3′ splice sites and the branch sequence are recognized by components of the splicing apparatus that assemble to form a large complex. This complex brings the 5′ and 3′ splice sites together before any reaction occurs, which explains why a deficiency in any one of the sites may prevent the reaction from initiating. The complex assembles sequentially on the pre-mRNA and passes through several “presplicing complexes” before forming the final, active complex, which is called the spliceosome. Splicing occurs only after all the components have assembled.
The splicing apparatus contains both proteins and RNAs (in addition to the pre-mRNA). The RNAs take the form of small molecules that exist as ribonucleoprotein particles. Both the nucleus and cytoplasm of eukaryotic cells contain many discrete small RNA types. They range in size from 100 to 300 bases in multicellular eukaryotes and extend in length to about 1,000 bases in yeast. They vary considerably in abundance, from 105 to 106 molecules per cell to concentrations too low to be detected directly.
Those restricted to the nucleus are called small nuclear RNAs (snRNAs); those found in the cytoplasm are called small cytoplasmic RNAs (scRNAs). In their natural state, they exist as ribonucleoprotein particles (snRNPs and scRNPs). Colloquially, they are sometimes known as snurps and scyrps, respectively.
Another class of small RNAs found in the nucleolus, called smallnucleol ar RNAs (snoRNAs), are involved in processing ribosomal RNA .
The snRNPs involved in splicing, together with many additional proteins, form the spliceosome. Isolated from the in vitro splicing systems, it comprises a 50S to 60S ribonucleoprotein particle. The spliceosome may be formed in stages as the snRNPs join, proceeding through several presplicing complexes. The spliceosome is a large body, greater in mass than the ribosome.
FIGURE 1 summarizes the components of the spliceosome. The five snRNAs account for more than a quarter of its mass; together with their 41 associated proteins, they account for almost half of its mass. Some 70 other proteins found in the spliceosome are described as splicing factors. They include proteins required for assembly of the spliceosome, proteins required for it to bind to the RNA substrate, and proteins involved in constructing an RNA-based center for transesterification reactions. In addition to these proteins, another approximately 30 proteins associated with the spliceosome are believed to be acting at other stages of gene expression, which suggests splicing may be connected to other steps in gene expression (see the section later in this chapter titled Splicing Is Temporally and Functionally Coupled with Multiple Steps in Gene Expression).
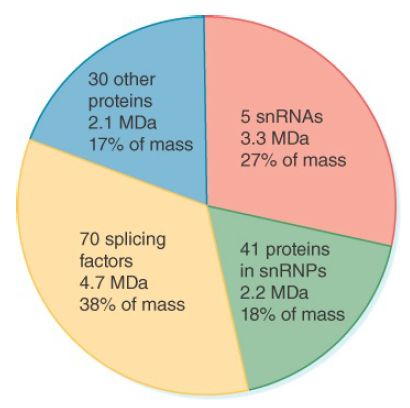
FIGURE 1. The spliceosome is approximately 12 megadaltons (MDa). Five snRNPs account for almost half of the mass. The remaining proteins include known splicing factors, as well as proteins that are involved in other stages of gene expression.
The spliceosome forms on the intact precursor RNA and passes through an intermediate state in which it contains the individual 5′ exon linear molecule and the right-lariat intron–exon. Little spliced product is found in the complex, which suggests that it is usually released immediately following the cleavage of the 3′ site and ligation of the exons.
We may think of the snRNP particles as being involved in building the structure of the spliceosome. Like the ribosome, the spliceosome depends on RNA–RNA interactions as well as protein–RNA and protein–protein interactions. Some of the reactions involving the snRNPs require their RNAs to base pair directly with sequences in the RNA being spliced; other reactions require recognition between snRNPs or between their proteins and other components of the spliceosome.
The importance of snRNA molecules can be tested directly in yeast by inducing mutations in their genes or in in vitro splicing reactions by targeted degradation of individual snRNAs in the nuclear extract.
Inactivation of five snRNAs, individually or in combination, prevents splicing. All of the snRNAs involved in splicing can be recognized in conserved forms in all eukaryotes, including plants. The corresponding RNAs in yeast are often rather larger, but conserved regions include features that are similar to the snRNAs of multicellular eukaryotes.
The snRNPs involved in splicing are U1, U2, U5, U4, and U6. They are named according to the snRNAs that are present. Each snRNP contains a single snRNA and several (fewer than 20) proteins. The U4 and U6 snRNPs are usually found together as a di-snRNP (U4/U6) particle. A common structural core for each snRNP consists of a group of eight proteins, all of which are recognized by an autoimmune antiserum called anti-Sm; conserved sequences in the proteins form the target for the antibodies. The other proteins in each snRNP are unique to it. The Sm proteins bind to the conserved sequence A/GAU Gpu, which is present in all snRNAs except U6. The U6 snRNP instead contains a set of Sm-like (Lsm) proteins.
Some of the proteins in the snRNPs may be involved directly in splicing; others may be required in structural roles or just for assembly or interactions between the snRNP particles. About onethird of the proteins involved in splicing are components of the snRNPs. Increasing evidence for a direct role of RNA in the splicing reaction suggests that relatively few of the splicing factors play a direct role in catalysis; most splicing factors may therefore provide structural or assembly roles in the spliceosome.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)