النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
In vitro DNA Replication
المؤلف:
Robert Schleif
المصدر:
Genetics and Molecular Biology
الجزء والصفحة:
2nd Edition , p60-62
2025-02-27
639
The mere incorporation of radiolabeled nucleotides into polymers of DNA is far from the complete biological process of DNA replication. The initial experiments seeking polymerization activities from cell extracts used nicked and gapped DNA as a template. This yielded polymerases capable of elongating DNA, but did not provide an assay for any of the DNA initiating components. To seek the cellular machinery necessary for initiating replication, DNA templates were required that contained sequences specifying origins of replication. The most convenient source of such origins was small DNA phages since each molecule must contain an origin. The results of experiments with several different phage templates revealed the astounding fact that the proteins required for initiating replication varied from one DNA origin to another. At first it was not possible to discern the biochemical principles underlying initiation of replication. Therefore, when DNA cloning became possible, attention turned to a replication origin of greater generality and importance, the origin of replication of the E. coli chromosome. Later, when it became possible to work with animal viruses and to isolate and study replication origins from eukaryotic cells, these also were studied.
Kornberg and his collaborators were able to find conditions in which a cell extract prepared from E. coli could replicate DNA from the E. coli origin, oriC. Such an extract undoubtedly possessed many different proteins acting in concert to replicate the DNA. Once this step was working, it was then possible to seek to identify specific proteins involved in the reaction. Geneticists assisted this difficult step through their isolation of temperature-sensitive mutations that blocked DNA synthesis in growing cells. For example, extracts prepared from cells with a temperature-sensitive dnaA mutation were inactive. This, of course, is a biochemist’s dream, for it provides a specific assay for the DnaA protein. Extracts prepared from wild-type cells can supplement extracts prepared from temperature-sensitive dnaA mutant cells. This supplementation results from the wild-type DnaA protein in the wild type extract. Next, the wild-type extract can be fractionated and the in vitro complementation assay detects which fraction contains the DnaA protein. With such an assay the DnaA protein was purified.
The strategy used with the DnaA protein is straightforward, and it can be used to purify proteins by making use of any replication mutant whose cell extracts are inactive. The work required for this approach is immense, and therefore it helps to try to guess proteins required for replication and to add these as purified components. If the assay doesn’t replicate DNA, cell extracts are added and the resulting activity can be used to guide purification of the remaining components. Ultimately, the following components were identified as necessary for in vitro replication from oriC: DnaA protein, DnaB protein, DnaC protein, DnaG protein, DNA polymerase III holoenzyme, DNA gyrase, single-stranded binding protein, and ATP. Analogous experiments using replication origins from animal viruses have also permitted the detection, purification and study of the complete set of proteins necessary for their activity.
In vitro initiation and synthesis reactions have permitted the replication process at the E. coli oriC to be dissected into several steps. Initiation begins with 20 to 40 molecules of DnaA protein binding to four sites in the 260 base pair oriC region (Fig. 1). The complex of DnaA and oriC contains the DNA wrapped around the outside and the protein in the middle. The large number of protein molecules required for the first step of initiation makes the step critically dependent on the concentration of the protein. Such a step is ideal for tight regulation of the initiation process. In the second step, DnaB plus DnaC bind. DnaB possesses a helicase activity which separates the DNA strands with the consumption of energy. DnaA binds to three additional sites in oriC and several regions of single-stranded DNA are generated. Finally, DnaG protein lays down RNA primers that are used by the pol III holoenzyme.

Fig1. The initiation of DNA replication from OriC by the combined activities of DnaA, DnaB, DnaC, and Dna pol III.
During the elongation process the single-stranded binding protein SSB binds to single-stranded regions opened by the helicase activities of DnaB (Fig. 2). DNA gyrase also is required for elongation. It untwists the rotations that are generated ahead of the moving replication fork. In vivo the twists that are generated behind the moving replication fork are removed by topisomerase I.
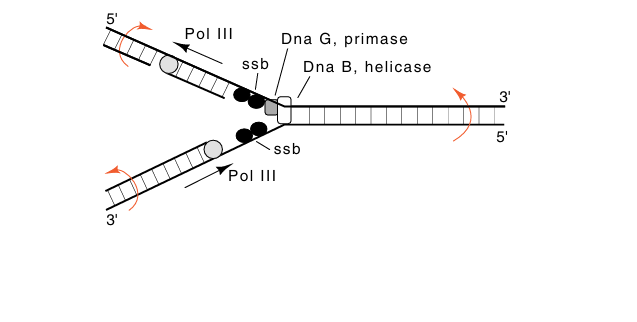
Fig2. The activities in the vicinity of a DNA replication fork. Also shown are the rotations generated by movement of a replication fork.
Several other proteins are also involved with replication. The histone- like protein, HU, seems to assist the process, but its mechanism is unknown. RNAse H digests the RNA from RNA-DNA hybrids left over from transcription and prevents initiation from points other than the origin.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)